Enzymes are complex biomolecules (proteins) that catalyse reactions in the cell. How the first enzymes evolved is a billion of years old mystery. To study early enzyme evolution, ELSI scientists looked for differences and similarities between some of the most ancient enzyme families. Their results highlight the hypotheses of the Late Prof. Dan S. Tawfik, who argued that phosphate binding loops – short segments of an enzyme that interact with phosphate – gave rise to several of the most important enzyme families. In this way, scientists can better understand modern biological structures’ history and simple origins.
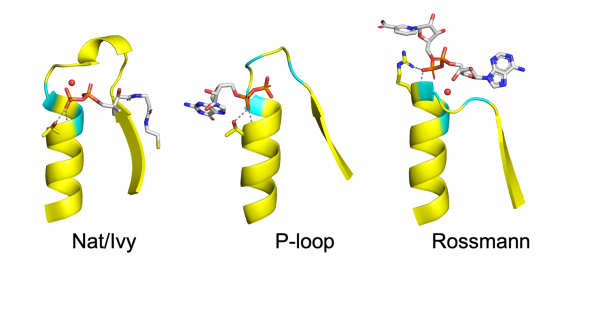
Figure 1. Common Phosphate Binding Loops (PBLs) in biology. PBLs (yellow and cyan) bind to cofactors, and especially phosphate (orange). Credit: Adapted from Liam M. Longo et al. Protein Science, 2022
Prof. Tawfik was a pioneer of protein evolution and a central figure in the invention of laboratory evolution techniques. Tragedy struck on May 4 2021, when Prof. Tawfik died in a rock-climbing accident in Croatia. To honour his memory, the academic journal Protein Science arranged for a special memorial issue and invited ELSI Specially-Appointed Associate Professor Liam M. Longo – a former student of the Tawfik Lab — to contribute an article. Before his death, Prof. Tawfik was captivated by a molecular structure called a phosphate binding loop or PBL. These structures are present in every single organism on Earth, from bacteria to humans, and are essential for enzyme function. Without PBLs, life as we know it could not exist. What better way to honor Prof. Tawfik’s memory than to celebrate (and interrogate) his research ideas about PBLs! To perform this study, Prof. Longo teamed up with Earth-Life Science Course Graduate Student Hayate Hirai and ELSI Associate Professor Shawn E. McGlynn.
The enzyme family that the scientists considered is called Nat/Ivy. This enzyme family is important because it is associated with diverse biological processes. For example, bacteria use Nat/Ivy enzymes to protect themselves from toxins that they produce to kill competing cells in the environment. At the heart of the Nat/Ivy structure is a PBL (Figure 1) that binds to a special metabolite called a cofactor (in this case, the cofactor Coenzyme A). Prof. Tawfik argued that PBLs with a simple structure, like those in Figure 1, may have been the origins of several different protein families. And in the case of Nat/Ivy, the PLB was very similar to PBLs in other proteins.
Although the authors cannot say for sure, the current evidence suggests that other PBLs are unrelated to that of Nat/Ivy – that is, they do not share a common ancestor. In the field of protein evolution, when two structures look similar to each other but are not evolutionarily related, it is called convergence. Convergent evolution suggests that the common features of the PBL are particularly good starting points for protein evolution because they appear time and again in biology.
One of the central challenges in the field of protein evolution is understanding how simple structures from ancient biology became the intricate molecular machines that are found in organisms on Earth today. Particularly mysterious is what the most ancient protein forms looked like. The Nat/Ivy example provides even more support for Prof. Tawfik’s guiding hypothesis: That simple PBLs may have been an important starting point for the evolution of many different modern enzymes (Figure 2). In this way, we can better understand the biological march towards increasing enzyme complexity over time.
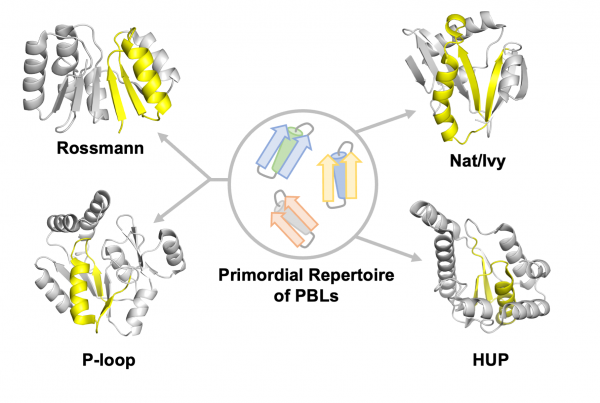
Figure 2. Prof. Tawfik’s Theory: A primordial repertoire of PBLs gave rise to many important modern enzymes (for example, Nat/Ivy, Rossmann, P-loop, and HUP). Credit: Adapted from Liam M. Longo et al. Protein Science, 2022
| 掲載誌 | Protein Science |
| 論文タイトル | An Evolutionary History of the CoA-Binding Protein Nat/Ivy |
| 著者 | Liam M. Longo1,2, Hayate Hirai1,3, Shawn E. McGlynn1,2, 3,4 |
| 所属 | 1. Earth-Life Science Institute, Tokyo Institute of Technology, Tokyo, Japan 2. Blue Marble Space Institute of Science, Seattle, Washington, USA 3. School of Life Sciences and Technology, Tokyo Institute of Technology, Tokyo, Japan 4. Biofunctional Catalyst Research 82 Team, RIKEN Center for Sustainable Resource Science, Saitama, Japan |
| DOI | 10.1002/pro.4463 |
| 出版日 | 3 October 2022 |
