Proteins twist themselves into intricate structures made up of thousands of atoms. These structures, called folds, are essential for biological function. One of the mysteries of biology is where folds came from in the first place. New results demonstrate that fold emergence can be facilitated by a unique environment: tiny droplets formed by a network of interacting peptides and RNA. Inside droplets, peptides can assemble to form complex structures that are then elaborated on by evolutionary processes. Droplets are an exciting avenue for the origin of life research, and these results suggest that their significance is even greater than previously known.
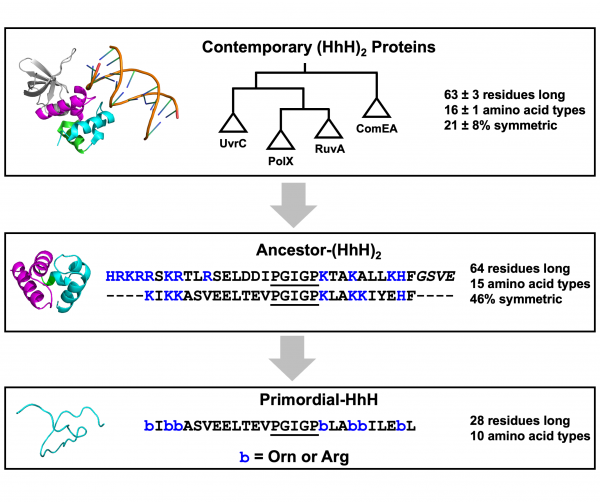
Modern biology hinges on the action of proteins, which are the primary workhorses of the cell. To achieve their functions, proteins must adopt intricate structures called folds (Figure 1). And while there have been major advancements in predicting the shape of a protein fold, the evolutionary origins of most folds remain a mystery. Specially Appointed Associate Professor Liam Longo (ELSI), Prof. Daniella Goldfarb (Weizmann Institute of Science), and Prof. Norman Metanis (Hebrew University of Jerusalem) had a hypothesis: Perhaps folds emerged inside droplets formed by mixing RNA and simple peptides. Indeed, droplets such as this have received considerable attention by origins of life research, starting with Oparin himself!
Since the history of protein evolution dates back nearly 4 billion years and cannot be rewound and replayed, how did the researchers test their ideas? First, they needed to choose a model system. To do this, they identified a protein family that is present in all organisms on earth, suggesting that it is one of the most ancient protein families. With all known sequences of this protein family in hand, the researchers could “resurrect” the ancestral protein using mathematical evolution models. Finally, the resulting “ancestor sequence” was further simplified – for example, by removing complex chemical groups that emerged only later in evolution (Figure 1).
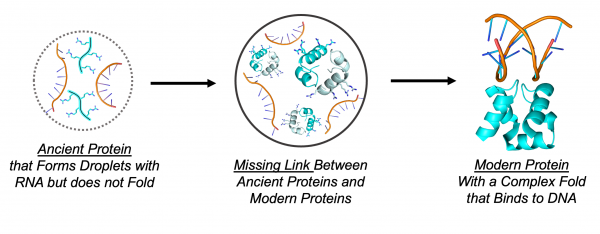
With a model primordial protein in hand, the researchers were ready to test the properties of this protein inside droplets that form when mixed with RNA. The internal structure of protein-RNA droplets is not well understood, so it was unknown whether the protein would adopt a random, spaghetti-like structure or whether it could fold up into a compact structure. To answer this question, the researchers turned to a technique called electron paramagnetic resonance, which can report on the conformational dynamics of proteins inside a droplet by watching the behavior of unpaired elections.
The results were striking: outside the droplets, the primordial protein was largely unfolded. This observation is common for very simple proteins because it is difficult to encode complex structures with very simple sequences. But when the researchers peered into the droplets, strong signatures of folding were observed. The unique conditions within a droplet – caused by the very high concentrations of RNA and peptide – promoted the attainment of a protein fold. In this way, previously inaccessible protein configurations could have been made accessible to nascent life.
One of the great challenges of molecular evolution is understanding how simple structures transformed into complex structures. The discovery that droplets made of RNA and protein can serve as a testing ground for new protein structures fills in a key intermediate stage and teaches researchers how biology may have discovered the first protein folds.
| Journal | Journal of the American Chemical Society |
| Tile of the paper | Peptide-RNA Coacervates as a Cradle for the Evolution of Folded Domains |
| Authors | Manas Seal1, Orit Weil-Ktorza2, Dragana Despotović3, Dan S. Tawfik3, Yaakov Levy4, Norman Metanis2,5,6, Liam M. Longo7,8,*, and Daniella Goldfarb1,* |
| Affiliations | 1.Department of Chemical and Biological Physics, Weizmann Institute of Science, Rehovot 7610001, Israel 2.Institute of Chemistry, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel 3.Department of Biomolecular Science, Weizmann Institute of Science, Rehovot 7610001, Israel 4.Department of Chemical and Structural Biology, Weizmann Institute of Science, Rehovot 7610001, Israel 5.Casali Center for Applied Chemistry, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel 6.The Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel 7.Earth-Life Science Institute, Tokyo Institute of Technology, Tokyo 152-8550, Japan 8.Blue Marble Space Institute of Science, Seattle, Washington 98104, United States |
| DOI | 10.1021/jacs.2c03819 |
| Online published date | 29 July 2022 |
