RNA-binding proteins play critical roles in various cellular processes, including DNA repair, post-transcriptional modification, and cancer progression. While nature provides a number of naturally occurring RNA-binding proteins, the design of de novo RNA-binding peptides remains challenging. Researchers at ELSI present a novel platform for the discovery of novel single-strand RNA-binding peptides that offer promising avenues for the regulation of RNA functions.
Credit: Nishikawa et.al., Biomacromolecules 2023.
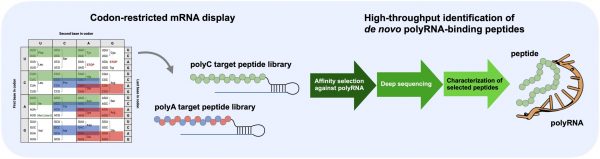
The development of novel RNA-binding proteins (RBPs) not only expands the toolbox of biotechnology, but also contributes to the elucidation of fundamental mechanisms governing nucleic acid-protein interactions. This knowledge could serve as a guide for the future design of artificial RBPs. The research team consisting, ELSI Professors Naohiro Terasaka and Kosuke Fujishima; ELSI Graduate Student Shota Nishikawa; and a collaborator from the University of Tokyo, extensively explored peptide sequence spaces that allow single-strand RNA binding by combining a codon-restricted DNA library with an mRNA display method.
The mRNA display method physically links peptides and their genetic information (mRNA), enabling high-throughput exploration of functional peptides in vitro. In this study, two types of simple RNA molecules (poly(A) RNA and poly(C) RNA) were selected as target molecules. To prevent base pairing between the target RNA and the mRNA portion, the team restricted the use of codons in the peptide-coding mRNA. Starting with a peptide library of ~1012 sequence diversity (33 amino acid length), the research team performed a total of seven rounds of selection (Figure 1).
After the in vitro selection, next-generation sequencing analysis was performed to reveal the sequence features that are important for RNA-binding capability within the selected de novo RNA-binding peptide sequences (Figure 2). As a result, the researchers found four new single-stranded RNA-binding peptides with enriched asparagine (N) and glutamine (Q) containing motifs important for RNA recognition. In addition, the newly identified poly(C) RNA-binding peptide (CP1) showed a strong affinity towards natural cytosine-rich oncogenic RNA motifs (Figure 3).
The method established in this study serves as a powerful tool for identifying novel RNA-binding peptides. In addition, this approach can be applied to explore the nucleic acid binding potential of peptides with a limited set of amino acids related to origin of life research as well as to explore novel peptide aptamers for future peptide drug discovery.
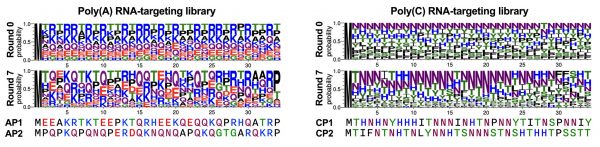
The sequence logo represents the abundance of amino acids at each position before and after selection of the random peptide library. After selection, significant enrichment of glutamine (Q) (for the poly(A) RNA-targeting peptide library) and asparagine (N) (for the poly(A) RNA-targeting peptide library) was observed. Credit: Nishikawa et.al., Biomacromolecules 2023.

| Journal | Biomacromolecules |
| Title of the paper | De novo single-stranded RNA-binding peptides discovered by codon-restricted mRNA display |
| Authors | Shota Nishikawa1,2 Hidenori Watanabe1, Naohiro Terasaka1, Takayuki Katoh3, and Kosuke Fujishima1,4 |
| Affiliations | 1. Earth-Life Science Institute, Tokyo Institute of Technology, Ookayama, Meguro-ku, Tokyo 152-8550, Japan 2. School of Life Science and Technology, Tokyo Institute of Technology, Ookayama, Meguro-ku, Tokyo 152-8550, Japan 3. Department of Chemistry, Graduate School of Science, The University of Tokyo, Bunkyo-ku, Tokyo 113-0033, Japan 4. Graduate School of Media and Governance, Keio University, Fujisawa 252-0882, Japan |
| DOI | 10.1021/acs.biomac.3c01024 |
| Online published date | 5 December 2023 |
