[Press Release]
Modern eukaryotic cells have proteins that enable chromosome segregation during cell division, new discoveries shed light on their origin in simpler prokaryotic organisms.
All modern organisms fall into two classes, eukaryotes and prokaryotes. Eukaryotes (from the Greek meaning “true kernel”) have a cell nucleus that harbours most of the cell’s genetic information and includes organisms such as humans, plants and fungi. In prokaryotes, the cell’s contents, including its genetic material, are diffusely distributed. Eukaryotes typically have much larger genomes, which is generally thought to relate to their greater complexity: indeed, all multicellular organisms are eukaryotes. Prokaryotes, however, not to be outclassed, comprise the bulk of Earth’s biomass. Generally speaking, prokaryotes seem to approach the process of living by making copies of themselves as quickly as they can, while eukaryotes survive by being highly specialized at making copies of themselves in particular ways.
A new study by a team of scientists including researchers at the Earth-Life Science Institute (ELSI) at Tokyo Institute of Technology, Research Institute for Interdisciplinary Science (RIIS) at Okayama University and Nagoya University may have identified a key intermediate in the transition from prokaryotes to eukaryotes.
Most prokaryotes have single circular genomes and reproduce asexually, which means when the time comes for their cells to divide, they simply have to copy their circular genomes and make sure one circular copy ends up in each daughter cell. In contrast, eukaryotes typically have multiple linear chromosomal genomes and often reproduce sexually, meaning they have to make sure an appropriate set of multiple copied chromosomes ends up in each daughter cell. This is a much more complicated process, and it remains unclear how the precise sorting required for this process arose from the simpler system used by prokaryotes.
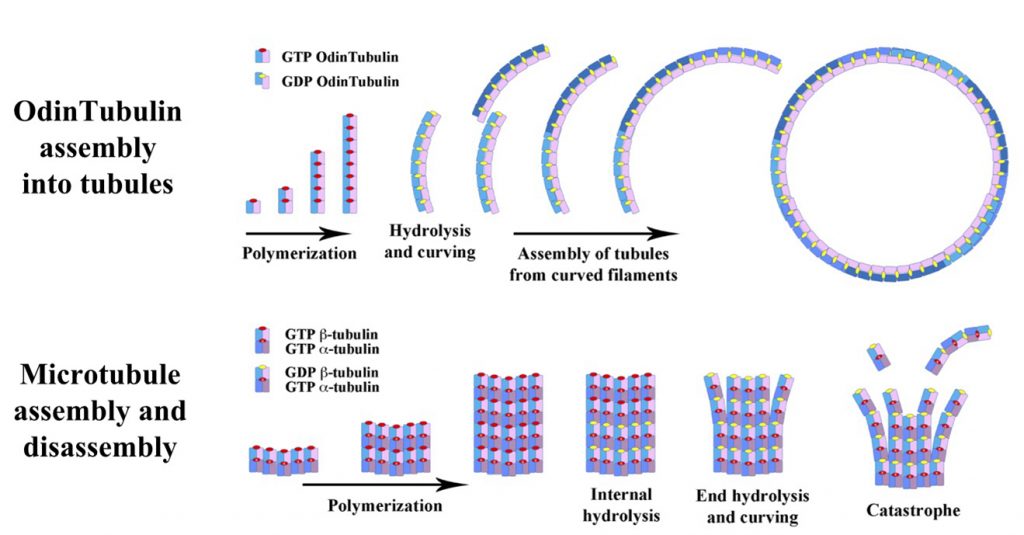
In prokaryotes, typically, the circular chromosome becomes attached to the cell membrane, and then as the cell grows and begins to separate into two cells, this attachment assures that one copy will end up in each daughter cell. This process is much more complex in eukaryotes. In eukaryotes, a complex protein scaffolding forms, mainly based on the protein tubulin. Tubulin forms long fibres, which help draw the copied chromosomes towards the poles of the dividing cell. Failures of this process, called nondisjunction, result in unequal numbers of chromosomes being present in daughter cells, which gives rise to disorders collectively known as aneuploidy. In humans, this leads to numerous recognizable birth defects, perhaps most notably Down’s Syndrome.
How the processes arose that allowed eukaryotic cells to precisely segregate chromosomes have been a mystery for some time. Prokaryotes produce a protein similar to tubulin, but rather than helping move chromosomes about, it helps pinch off the daughter cell from the parent. This protein is known as FtsZ.
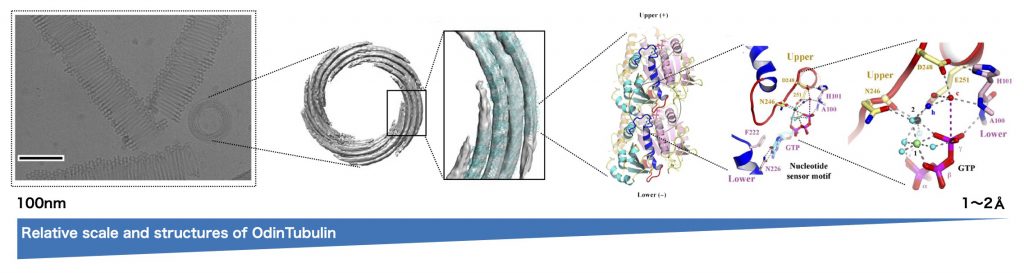
When prokaryotes and eukaryotes split is yet another evolutionary mystery, this split is generally believed to have occurred ~ 1-2 billion years ago, but recently a group of organisms that appear to be evolutionarily intermediate has been identified. These organisms are known as the Asgard archaea, whose name references Norse creation myths. A team of scientists, including Caner Akil and Kosuke Fujishima of ELSI, has now identified a protein similar to eukaryotic tubulin in the genome of an Asgard species isolated from the hot spring waters of Yellowstone National Park.
“These new asgard archaeal proteins, which the scientists have named OdinTubulin, in another shout-out to the Norse pantheon, are similar to both eukaryotic tubulins and prokaryotic FtsZ proteins,” says Samson Ali, Nagoya University and Okayama University. Linh T. Tran, Okayama University, adds that “OdinTubulin may therefore represent an evolutionary intermediate between prokaryotic FtsZ and eukaryotic microtubule-forming tubulins.”
Reference:
Caner Akıl1,2†, Samson Ali1,3†, Linh T. Tran1†, Jérémie Gaillard4, Wenfei Li5, Kenichi Hayashida6, Mika Hirose7, Takayuki Kato7, Atsunori Oshima6,8,9, Kosuke Fujishima2,10, Laurent Blanchoin4,11, Akihiro Narita3*, Robert C. Robinson1,12*, Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution, Science Advances, DOI: 10.1126/sciadv.abm2225
1. Research Institute for Interdisciplinary Science, Okayama University, Okayama 700-8530, Japan.
2. Tokyo Institute of Technology, Earth-Life Science Institute (ELSI), Tokyo 152-8551, Japan.
3. Division of Biological Science, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan.
4. University of Grenoble-Alpes, CEA, CNRS, INRA, Interdisciplinary Research Institute of Grenoble, Laboratoire de Physiologie Cellulaire & Végétale, CytoMorpho Lab, 38054 Grenoble, France.
5. National Laboratory of Solid State Microstructure, Department of Physics, Collaborative Innovation Center of Advanced Microstructures, Nanjing University, 210093 Nanjing, China.
6. Cellular and Structural Physiology Institute (CeSPI), Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan.
7. Institute for Protein Research, Osaka University, Osaka 565-0871, Japan.
8. Department of Basic Medicinal Sciences, Graduate School of Pharmaceutical Sciences, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan.
9. Institute for Glyco-core Research (iGCORE), Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8601, Japan.
10. Graduate School of Media and Governance, Keio University, Fujisawa 252-0882, Japan.
11. Université de Paris, INSERM, CEA, Institut de Recherche Saint Louis, U 976, CytoMorpho Lab, 75010 Paris, France.
12. School of Biomolecular Science and Engineering (BSE), Vidyasirimedhi Institute of Science and Technology (VISTEC), Rayong 21210, Thailand.
*Corresponding authors’ email addresses: br.okayama.u@gmail.com (R.C.R.); narita.akihiro@f.mbox.nagoya-u.ac.jp (A.N.)
Contacts:
Thilina Heenatigala
Director of Communications
Earth-Life Science Institute (ELSI),
Tokyo Institute of Technology
E-mail: thilinah@elsi.jp
Tel: +81-3-5734-3163 / +81-3-5734-3416
Kosuke Fujishima
Associate Professor
Earth-Life Science Institute (ELSI),
Tokyo Institute of Technology
E-mail: fuji@elsi.jp
Tel: +81-3-5734-3199
