[This is a Joint Press Release with Weizmann Institute of Science and the Hebrew University of Jerusalem ]
Researchers found a rare example of an ancient protein that can still function in a molecular mirror world – offering new clues about the evolutionary history of proteins that bind to nucleic acids. But many mysteries remain.
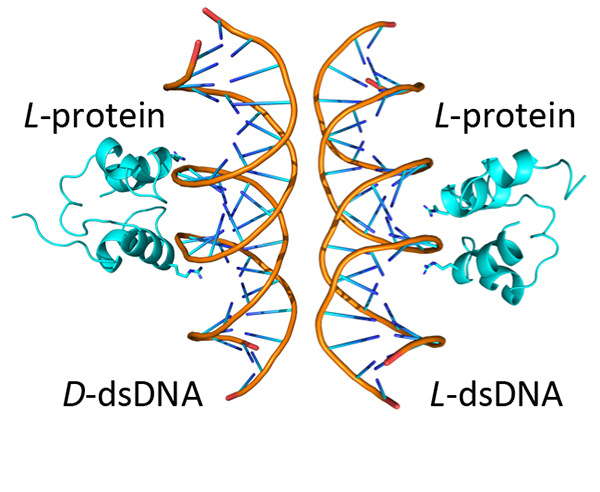
A simplified (HhH)2-Fold protein binding to natural and mirror-image dsDNA. The binding surface between the two binding modes is partially overlapping Credit: Longo, L. M. et al., Angewandte Chemie International Edition (2025).
A new study led by researchers from the Earth-Life Science Institute (ELSI) at Institute of Science Tokyo, in collaboration with the Hebrew University of Jerusalem and the Weizmann Institute of Science, found that an ancient protein motif that binds to nucleic acids is functionally “ambidextrous.” This means that the motif can interact with both natural and mirror-image nucleic acids, an occurrence that has never before been reported for nucleic acid binding. The study provides a unique snapshot of protein evolution and raises intriguing questions about the origins and history of life on Earth.
Like the human hand, many molecules exist in “right-handed” and “left-handed” forms that are mirror images of each other. Life on Earth has a strong preference for molecular handedness: proteins are made up of left-handed amino acids, while nucleic acids are made up of right-handed sugars. This phenomenon, called homochirality, is a fundamental feature of biology as we know it. With few exceptions, flipping the handedness of a complex molecule causes it to fall out of step with Earth life and to stop functioning. It is kind of like Alice stepping through the looking glass into a world quite unlike her own. This fact has led to a long-standing view that mirror-image proteins cannot perform their normal biological function.
New research led by Liam M. Longo, Specially Appointed Associate Professor and graduate student Tatsuya Corlett at ELSI, in collaboration with professor Norman Metanis and research scientist Orit Weil-Ktorza at the Hebrew University of Jerusalem and professor Yaakov (Koby) Levy, PhD. Student Segev Naveh-Tassa, and research scientists Yael Fridmann-Sirkis, Dragana Despotović, and Kesava Phaneendra Cherukuri at the Weizmann Institute of Science, has challenged this assumption. In the study, published in the journal Angewandte Chemie, the researchers reported that a primordial and widely conserved protein structure called the helix-hairpin-helix (HhH) motif is capable of functioning in both left- and right-handed forms. In its natural state, the HhH motif binds to DNA and RNA. Surprisingly, the researchers found that a chemically synthesised mirror image of the motif is also functional—a phenomenon the authors refer to as functional ambidexterity. In simple terms, the HhH motif is like a glove that fits snuggly on both hands. No such nucleic-acid binding protein has been reported before.
Double stranded DNA has a right-handed twist, so the reasonable expectation was that a mirror-image protein would be unable to bind. “I was looking at the motif – just playing around on the computer – and I suddenly thought: This motif can bind mirror-DNA!” said Longo. “It was a crazy idea,” said Metanis, a peptide chemist, ‘but the more we looked at the structure, the more we thought that maybe we were on to something.” To investigate whether the mirrored HhH motif-containing proteins bind to double stranded DNA, they synthesised the mirror-image protein and measured its binding in the lab. Sure enough, binding was detected.
But was binding of the mirror-image protein similar to the natural protein? To test this, the researchers analysed the kinetics of un-binding, as well as the effect of several mutations. And both approaches suggested surprising similarities. Encouraged by these results, Longo reached out to Levy, a computational chemist who could use molecular simulations to paint a detailed molecular picture of the binding process. What the researchers found surprised them: Although there were differences in how the natural and mirror-image proteins bound to DNA, similar regions of the protein were responsible for binding in each case. In other words, these two binding modes were linked to each other at the molecular level.
This study is an exciting first example of ambidextrous proteins binding to nucleic acids, and raises more questions than it answers. What forces caused this protein to be ambidextrous in the first place? “We still don’t know,” says Metanis. “Perhaps it reflects the need for this domain to bind multiple types of DNA structures, or to slide along the DNA molecule.” Or, perhaps it is pointing to an even greater evolutionary mystery: “The most intriguing explanation is that functional ambidexterity was once under selective pressure, perhaps due to ancient mirror-image life!” said Longo with a smile. But the authors caution that we are still a long ways away from being able to draw such provocative conclusions. For now, the hunt for other ambidextrous proteins begins!
Reference:
Orit Weil-Ktorza‡,1, Segev Naveh-Tassa‡,2, Yael Fridmann-Sirkis‡,3, Dragana Despotović‡,4,5, Kesava Phaneendra Cherukuri4, Tatsuya Corlett6, Yaakov Levy2,*, Norman Metanis1,*, Liam M. Longo6,7,*. Functional Ambidexterity of an Ancient Nucleic Acid-Binding Domain, Angewandte Chemie. DOI: https://doi.org/10.1002/anie.202505188
- Institute of Chemistry, The Center for Nanoscience and Nanotechnology, Casali Center of Applied Chemistry, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel
- Department of Chemical and Structural Biology, Weizmann Institute of Science, Rehovot 7610001, Israel
- Department of Life Sciences Core Facilities, Weizmann Institute of Science, Rehovot 7610001, Israel
- Department of Biomolecular Sciences, Weizmann Institute of Science, Rehovot 7610001, Israel
- Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade 11042, Serbia
- Earth-Life Science Institute, Institute of Science Tokyo, Tokyo 152-8550, Japan
- Department Blue Marble Space Institute of Science, Seattle, Washington 98104, USA
[‡] These authors contributed equally to this work.
*Corresponding author’s email: llongo@elsi.jp
Contacts:
Thilina Heenatigala
Director of Communications
Earth-Life Science Institute (ELSI),
Institute of Science Tokyo
E-mail: thilinah@elsi.jp
Tel: +81-3-5734-3163 / Fax: +81-3-5734-3416
Liam M. Longo
Specially Appointed Associate Professor
Earth-Life Science Institute (ELSI),
Institute of Science Tokyo
Email: llongo@elsi.jp
